GreenThumb Bud Count & Area Analysis
This blog was originally published on the GreenThumb IO Medium Leaf Area Analysis.
Intro
I gleamed my inspiration from PyImageSearch.com yet again. I’m basically counting bright green spots, by combining some of the tricks from the last post on Leaf Area Analysis.
Detecting multiple bright spots in an image with Python and OpenCV - PyImageSearch
Dependencies
In this docker image I have all of the dependencies for both Leaf Area and Bud Area including working copies and works in progress. We’re going to be using the same setup as with the Leaf Area notebook. OpenCV will be used for preprocessing the image and pixel selection by range. Matplotlib is used from rendering images and graphs. We take advantage of skimage.measure.label for taking measurements and label them. We take these components and use OpenCV to find and annotate the contours.
HSV Range Selector
open -a XQuartz
ip=$(ifconfig en0 | grep inet | awk '$1=="inet" {print $2}')
display_number=`ps -ef | grep "Xquartz :\d" | grep -v xinit | awk '{ print $9; }'`
/opt/X11/bin/xhost + $ip
docker run -it --rm -e DISPLAY=$ip:0 -v /tmp/.X11-unix:/tmp/.X11-unix tensorflow-notebook /bin/bash
range-detector --filter HSV --image work/test3.png --preview
I pulled the first few lines of the script below from this comment. It was need to run the range_detector used in this and the previous blog post.
Running GUI applications using Docker for Mac

You can checkout this GIF to get an idea of how I go about defining the HSV boundary values.Creating the initial HSV Mask
Creating the initial HSV Mask
Simple range mask like we did with the leaf detector
Preview the mask
We can easily mask the original image with OpenCV’s bitwise_and which takes the original image and the mask and outputs a masked image.
Cannabis Computer Vision
This is a side-by-side view of the HSV colorspace which the computer uses to isolate the buds and the masked preview. We use the numpy.vstack to vertically stack the images. I chose vstack for the gists here and hstack in the notebook, but it really is a matter of developer preference and aspect ratio.
Refining the Mask
We can refine the mask using the same binary threshold, erode, and dilate technique used on the Leaf Area. This will clean up the specs of frost leave from the actual Bud Area.
Before and After Cleaning Up the Contours
You can see the vstack` comparing the contours pre and post cleanup.
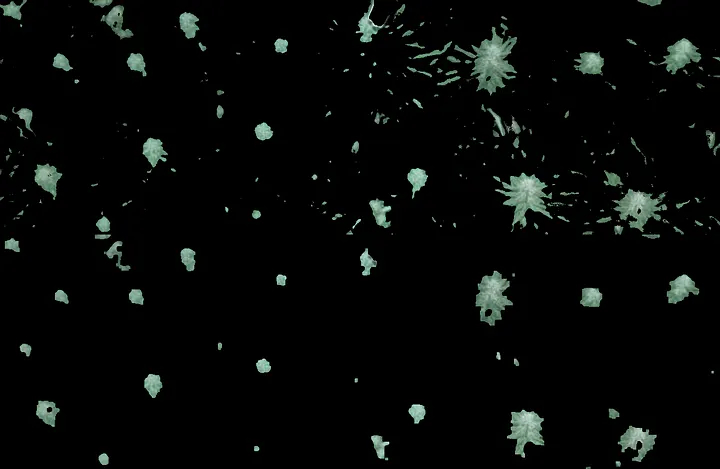 You can still see a few smaller contours that escape this sweep, but they’re technically smaller bud sites below the top layer of canopy. Either way we’ll filter them in the next pass below.
You can still see a few smaller contours that escape this sweep, but they’re technically smaller bud sites below the top layer of canopy. Either way we’ll filter them in the next pass below.
Enhance Mask with Labeled Components
Using skimage.measure.label we are further filtering out noise by ignoring blobs smaller than 100px. This means buds have to be this size or greater to get added to the labeled mask.
labels = measure.label(postprocessed_mask, neighbors=8, background=0)
mask = np.zeros(thresh.shape, dtype="uint8")
# loop over the unique components
for label in np.unique(labels):
# if this is the background label, ignore it
if label == 0:
continue
# otherwise, construct the label mask and count the
# number of pixels
labelMask = np.zeros(thresh.shape, dtype="uint8")
labelMask[labels == label] = 255
numPixels = cv2.countNonZero(labelMask)
# if the number of pixels in the component is sufficiently
# large, then add it to our mask of "large blobs"
if numPixels > 100:
mask = cv2.add(mask, labelMask)
Label the Buds
Now we have what we need to label the bud sites on the preview image.
 You can see the smaller spots, which actually appear to be smaller bud sites below the canopy are not labeled. We can remove or lower the threshold of 100px to further refine the camera’s calibration, but this is a great result for this stage.A Full Copy of the Notebook Below
Below is the full copy of the notebook. I also have a docker image tonsoffun/tensorflow-notebook which I’ve simply added OpenCV 3.3 via anaconda. This docker image supports both Bud and Leaf Area Analysis as well. Keep following along as I show you how we can use our OpenCV masks to train a Mask RCNN.
You can see the smaller spots, which actually appear to be smaller bud sites below the canopy are not labeled. We can remove or lower the threshold of 100px to further refine the camera’s calibration, but this is a great result for this stage.A Full Copy of the Notebook Below
Below is the full copy of the notebook. I also have a docker image tonsoffun/tensorflow-notebook which I’ve simply added OpenCV 3.3 via anaconda. This docker image supports both Bud and Leaf Area Analysis as well. Keep following along as I show you how we can use our OpenCV masks to train a Mask RCNN.
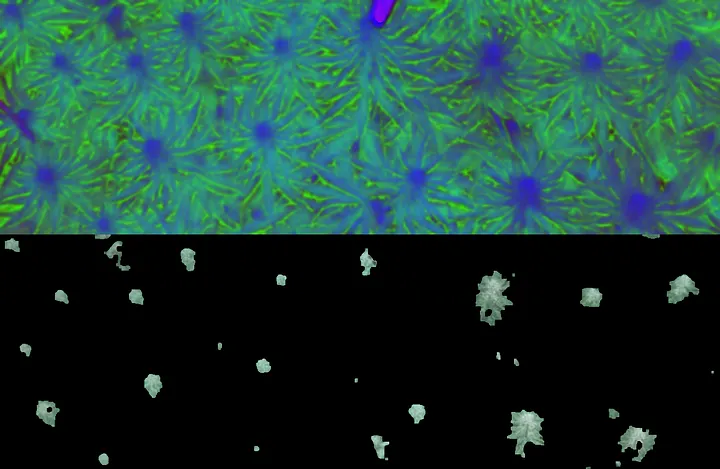
top: HSV Original, bottom: Final Mask Before Contour Filtering